Peptide Library Synthesis
Introduction
There is a growing need for highly efficient synthetic peptide library systems, due to recent developments in the discovery of novel drug targets. Based on the advantages of human genome projects and ongoing advancements in peptide delivery technology. Combinatorial chemistry has proven to be a useful technique for generating synthetic peptide libraries. It can produce a large number of distinct compounds simultaneously and screen them rapidly for a useful one. For example, combinatorial chemistry can be used to synthesize billions of different heptapeptides at the same time.
Enzymatic substrates and inhibitors or cell binding peptides can be screened by such peptide libraries. In contrast to the traditional synthetic approach, which involves working with one molecule at a time, combinatorial chemistry has been recognized as an important tool in the search for new drug candidates, catalysts, and materials.
Chemical Peptide Libraries
Merrifield invented solid-phase peptide synthesis (SPPS) in the 1960s, which serves as the foundation for the synthetic techniques used in peptide combinatorial libraries. In chemical approach, library synthesis occurs in two ways:
- when it is done on a solid support, an insoluble porous polymer resin, and the members are then cleaved for screening as free compounds.
- where the library is synthesized on a solid matrix and screened.
Parallel Synthesis
Parallel synthesis is a combinatorial synthesis, the single product is obtained in each different reaction flask. Alternatively, the procedure can be designed to mixtures of compounds are generated in each reaction vessel. Parallel synthesis can be performed in two different formats: one on a resin that is typically a cross-linked insoluble polymer bead, and another on a chemically functionalized paper pieces, which will be referred to as a membrane in this manuscript.
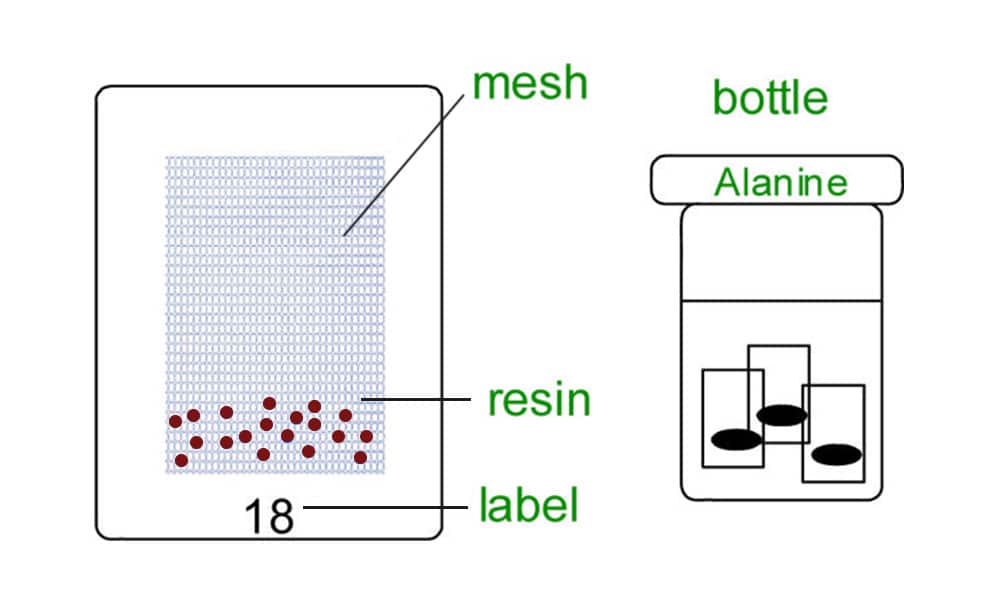
The teabag procedure is example of an early manual approach to parallel synthesis. Where the polymeric resin support is sealed in meshed containers with labels (called teabags). Subsequently, the teabags are placed in bottles that serve as the reaction vessels as shown in the figure below. Addition of the first amino acid to the resin in the case of peptide synthesis. In addition, different bottles have different amino acids. At this point, every teabag in a certain bottle has the same amino acid that is connected to the resin. All of the teabags from each bottle are placed in one container for deprotection and cleaning. After that, the teabags can be rearranged within the bottles to accommodate the addition of a second amino acid, combined again for deprotection and washing, redistributed to support the addition of the next amino acid, and so on.
SPOT synthesis is another parallel synthesis method. In this approach, a solution of active amino acids is positioned and delivered in the form of small drops to different points on a solid surface like a functionalized cellulose membrane or a glass slide, making a pattern of small dots. Based on traditional Fmoc-based peptide chemistry, peptide arrays are synthesized step-by-step. Automated SPOT synthesis with market instruments allows for the routine synthesis and screening of 600 peptides on a 150 cm2 membrane.
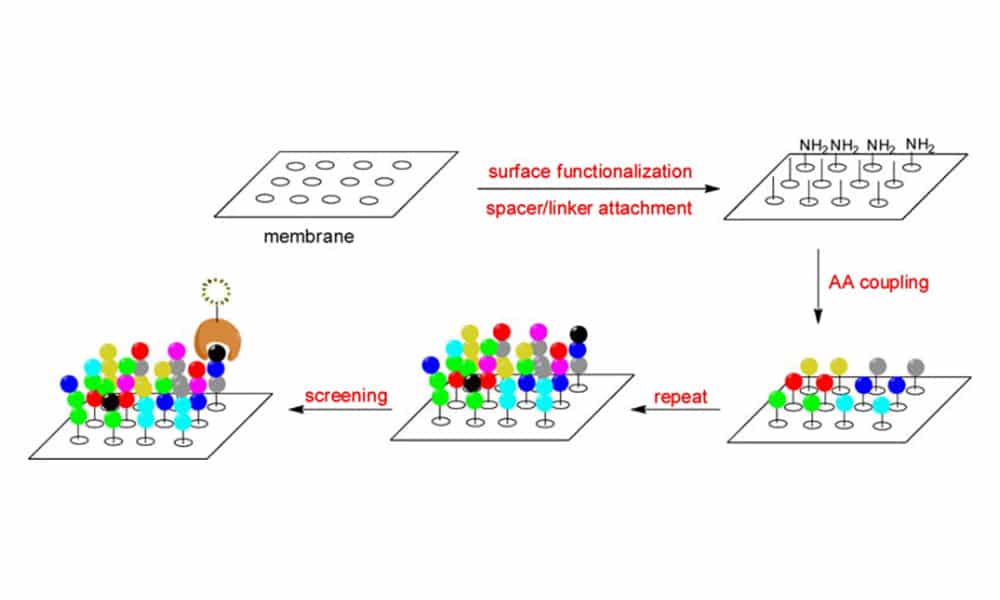
Split and Mix Synthesis
The split and mix approach, which uses peptide synthesis resin beads, can be utilized to create larger libraries. There are three steps to this strategy: (1) Resin beads are connected to every amino acid. (2) After combining the beads, they are divided into equal portions. The number of residues that will be connected determines the number of portions. Then a different amino acid is introduced to each component and reacts. (3) Carry out steps (1) and (2) once more until the peptides reach the required length. The figure below illustrates this by utilizing three amino acids (Gly, Ala, and Val) as building blocks to create a library of tripeptides.
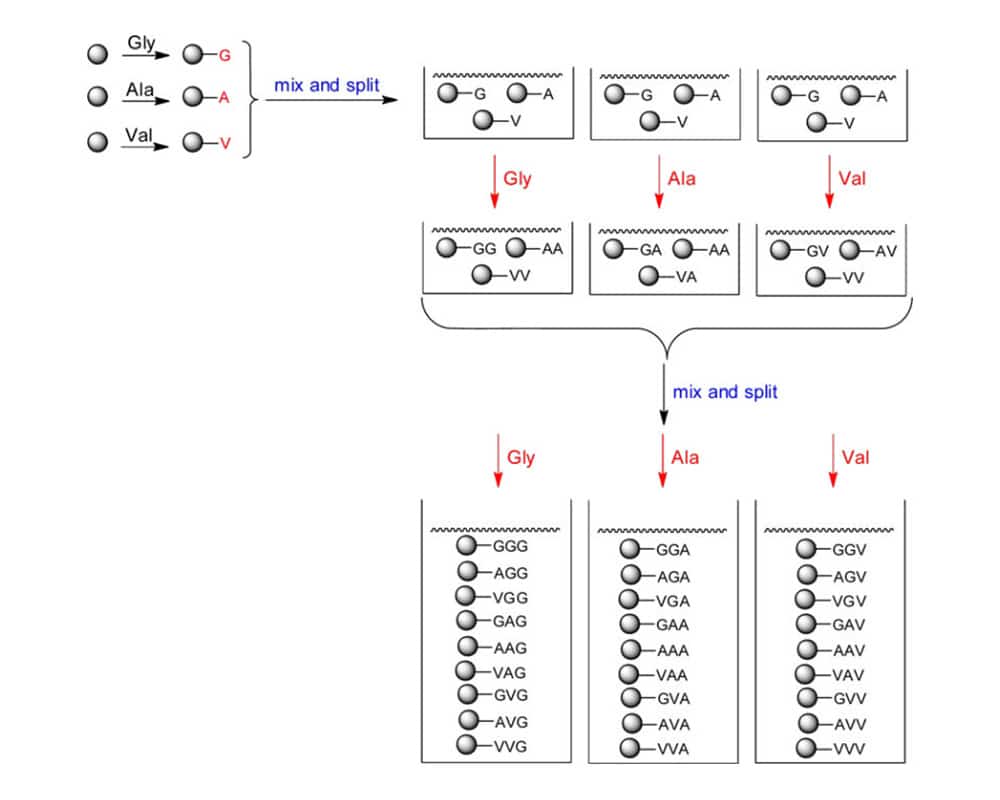
Following split and mix synthesis, every one of the 27 potential tripeptides will be created. It is an important feature of this method that each bead only contains one peptide sequence instead of a mixture. As a result, this method is also known as a one-bead-one-compound (OBOC) library. A library size of 205 = 3.2×106 members will be produced if a pentapeptide library made up of 20 distinct amino acids were prepared. The size is significantly larger than what parallel synthesis can generate. However, it is vital to emphasize that in this instance, the peptide sequence on a individual bead is unknown in advance. After the bead library has been screened, the peptide sequences of the selected beads must be determined by the following strategies: Edman degradation, partial Edman degradation mass spectrometry (PED/MS), or tandem mass spectrometry (MS/MS).
Reagent Mixture Synthesis
Compared to the split and mix method, the reagent mixture synthesis method is more convenient for large peptide libraries. In this method, building blocks are formed by coupling reaction involving all the amino acid reagent mixtures in one reaction chamber. Since the reaction rate of each amino acid reagent is different, it is necessary to calculate isokinetic ratios and use the corresponding amounts of each amino acid reagent in the coupling reactions to compensate. For example, Ostresh et al. calculated the isokinetic ratios of tert-butyloxycarbonyl (Boc)-amino acid to perform the reagent mixture synthesis. However, this method is not suitable for OBOC combinatorial library synthesis due to a mixture of peptide products may be contained by each single bead.
Reagent mixture synthesis as well as split and mix synthesis are beneficial for positional scanning in screening methods. Hexapeptide libraries for positional scanning is shown in the figure. Where ‘O’ represents a known residue in the mixture and is used for generating the library as a monomer; ‘X’ indicates an equimolar mixture of every amino acid that is utilized.
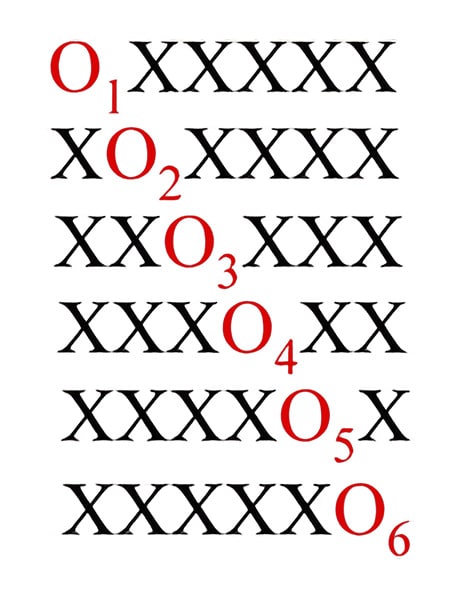
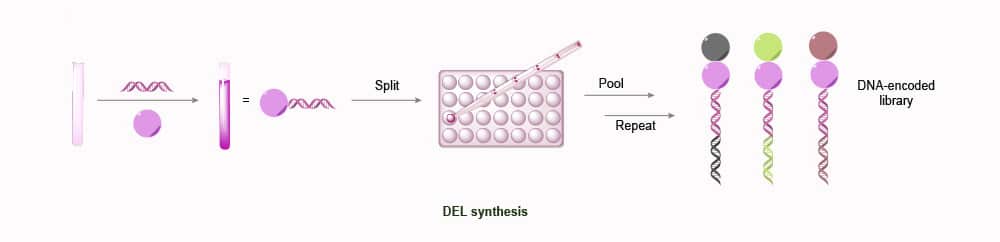
DNA-encoded Library (DEL) Platform
The DEL platform is cutting-edge technology for chemical combinatorial libraries. It works by library members are marked as double-stranded DNA barcodes via adapter modules, allowing retained compounds can be unambiguously identified at the end of the selection process. The “split-and-mix” approach is relied upon for library synthesis and tagging. Each synthesis cycle is encoded by ligating a short DNA tag, which identifies the added amino acid residue. Then affinity selection is used to analyze a pooled library, and the binders that were selected can be quickly identified by tag sequencing and PCR amplification. The DEL platform was used to identify a number of peptides, including CCR6, integrin ligands, and binders of carbonic anhydrase.
The following are several benefits that DEL provides for the finding of peptide ligands:
- Larger library sizes (i.e., diversity) enable for more in-depth sampling of the chemical space on a single screen.
- Parallel screening against multiple targets is compatible with the DEL platform, which can also perform activity-agnostic screening to identify “silent binders” and relatively low amounts of reagents are consumed.
- Individual screening hits can be identified. In addition, larger families, containing enriched combinations thereof and related building blocks can be also detected.
Additionally, the DEL platform has some limitations.
- Oligonucleotide tags may interfere with target-binder interactions, which may result in steric hindrance.
- The extent of possible chemical reactions can be limited by the barcode, and the properties of binders can also be affected.
- The assay takes longer to complete since it must be resynthesis to verify the subsequent target binding.
Conclusion
In this review, we describe the current SPPS technologies that are connected to combinatorial chemistry. The range of applications for synthesized peptide libraries is anticipated to expand continuously due to the tremendous advancements made in the field of SPPS during the past ten years. To guarantee your study success, QYAOBIO provides extensive Peptide Library services.